Attention: Confluence is not suitable for the storage of highly confidential data. Please ensure that any data classified as Highly Protected is stored using a more secure platform.
If you have any questions, please refer to the University's data classification guide or contact ict.askcyber@sydney.edu.au
3 Initiate jobs on Artemis
Example on how to run a DICOM to NIFTI conversion on Artemis.
In order to run on the Artemis, you need to have an Artemis account. For more information, please refer to below guide to logon to Artemis.
https://sydney.edu.au/research_support/hpc/access/index.shtml#newacct
For Artemis user guide, please refer to the University of Sydney Artemis User Guide
Step 1. Logon to Artemis
ddd-mbp:Deployment$ ssh unikey@hpc.sydney.edu.au
unikey@hpc.sydney.edu.au's password:
Last login: Wed May 9 13:53:09 2018 from vlan-2689-10-17-108-122.staff.wireless.sydney.edu.au
------------------------------------------------------------------------------
***************************** ARTEMIS 3.0 ******************************
------------------------------------------------------------------------------
*** !!! LOGIN NODES ARE FOR JOB SETUP & SUBMISSION ONLY !!! ***
------------------------------------------------------------------------------
Use Interactive flag (-I) or defaultQ or small-express queue for job testing
NO ACTUAL COMPUTE WORK IS TO BE CONDUCTED ON LOGIN NODES
Any sustained compute or multicore/memory intensive process on login nodes
will be subject to being killed without notice.
------------------------------------------------------------------------------
!!! MEMORY MUST BE SPECIFIED FOR ALL JOBS - use the mem= PBS directive !!!
------------------------------------------------------------------------------
Please refer to the Quick Start User Guide to get started with Artemis 2.0:
https://informatics.sydney.edu.au/services/artemis/
-------------------------------------------------------------------------------
Step 2. Download the XNAT data into Artemus directory into the folder. For data in big size, it is recommended to put in to the Artemis /scratch directory.
As the example is only a small set of data (32MB), we are putting it into the home folder on the Artemis Logging node.
[unikey@login2 ~]$ curl -u username "https://xnat.sydney.edu.au/data/archive/projects/PTV000/subjects/Haofei01/experiments/Haofei01_MR1/scans/4/resources/DICOM/files?format=zip" -o dicomfiles.zip
Enter host password for user 'unikey':
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 22.8M 0 22.8M 0 0 5827k 0 --:--:-- 0:00:04 --:--:-- 5994k
[unikey@login2 ~]$ ls -ltr
-rw-r--r-- 1 unikey linuxusers 23963682 May 11 09:22 dicomfiles.zip
[unikey@login2 ~]$ unzip dicomfiles.zip
Archive: dicomfiles.zip
extracting: Haofei01_MR1/scans/4-Ax_3D_T1/resources/DICOM/files/1.3.6.1.4.1.14301.53.4.1079983.1-4-134-4qu0ab.dcm
extracting: Haofei01_MR1/scans/4-Ax_3D_T1/resources/DICOM/files/1.3.6.1.4.1.14301.53.4.1079983.1-4-34-4qtzjm.dcm
...
[unikey@login2 ~]$ ls -ltr
total 23480
-rw-r--r-- 1 unikey linuxusers 23963682 May 11 09:22 dicomfiles.zip
drwxr-xr-x 3 unikey linuxusers 4096 May 11 09:24 Haofei01_MR1
Step 3. Create the Artemis Scripts
For information about Artemis Job Submission, monitoring and management, please refer to the information page about Artemis.
https://sydneyuni.atlassian.net/wiki/spaces/RC/pages/190284213/Job+Submission
https://sydneyuni.atlassian.net/wiki/spaces/RC/pages/213090348/Job+Monitoring+and+Management
[unikey@login2 ~]$ vi run.pbs
#!/bin/bash
#PBS -P SASTEST
#PBS -l select=1:ncpus=1:mem=1gb
#PBS -l walltime=1:00:00
#PBS -M emailemail@sydney.edu.au
#PBS -m abe
#PBS -j oe
module load dcm2niix
mkdir -p /home/unikey/Haofei01_MR1/scans/4-Ax_3D_T1/resources/NIFTI
dcm2niix -o /home/unikey/Haofei01_MR1/scans/4-Ax_3D_T1/resources/NIFTI/ /home/hfen0483/Haofei01_MR1/scans/4-Ax_3D_T1/resources/DICOM/files
Step 4. Run the Artemis Script
[unikey@login2 ~]$ qsub run.pbs
2238022.pbsserver
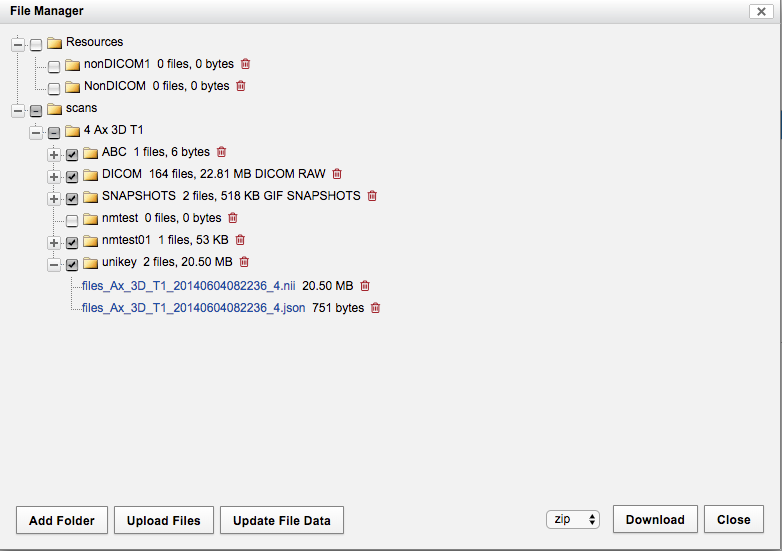
Step 5. Upload the result back to XNAT
[unikey@login2 ~]$ cd /home/unikey/Haofei01_MR1/scans/4-Ax_3D_T1/resources/NIFTI/
[unikey@login2 NIFTI]$ ls -ltr
total 21000
-rw-r--r-- 1 hfen0483 RDS-ICT-SASTEST-RW 21496160 May 11 09:29 files_Ax_3D_T1_20140604082236_4.nii
-rw-r--r-- 1 hfen0483 RDS-ICT-SASTEST-RW 751 May 11 09:29 files_Ax_3D_T1_20140604082236_4.json
Note, below command create the "unikey" folder and upload the file in one command in XNAT
[unikey@login2 NIFTI]$ curl -u username -X PUT "https://xnat.sydney.edu.au/data/archive/projects/PTV000/subjects/Haofei01/experiments/Haofei01_MR1/scans/4/resources/unikey/files" -F "file=@files_Ax_3D_T1_20140604082236_4.nii"
Enter host password for user 'unikey':
[unikey@login2 NIFTI]$ curl -u username -X PUT "https://xnat.sydney.edu.au/data/archive/projects/PTV000/subjects/Haofei01/experiments/Haofei01_MR1/scans/4/resources/unikey/files" -F "file=@files_Ax_3D_T1_20140604082236_4.json"
Enter host password for user 'unikey':